探索營養專家評選出的10種熱門飲食法排名、優缺點解析,以及它們如何幫助減重。
美國權威新聞雜誌《美國新聞與世界報導》最新公布的38種流行飲食法排名引發全球健康愛好者關注。這份榜單從營養價值、安全性、易執行性、減重效果與疾病預防等面向進行評估。我們特別整理榜單中表現最亮眼與最受爭議的飲食法,助您根據自身需求選擇最適合的飲食方式。

1. DASH飲食法 – 4.2/5
專為控制高血壓設計的DASH飲食法,憑藉護心功效與減重潛力榮登榜首。
特色:
- 強調低脂、低鈉食物
- 富含鉀、鎂、膳食纖維與不飽和脂肪
- 包含水果、蔬菜、全穀物、瘦肉蛋白與低脂乳製品
- 限制紅肉與高糖食品
專家觀點:雖營養均衡且易長期執行,但過量纖維與鉀攝取可能增加消化系統或腎臟負擔。

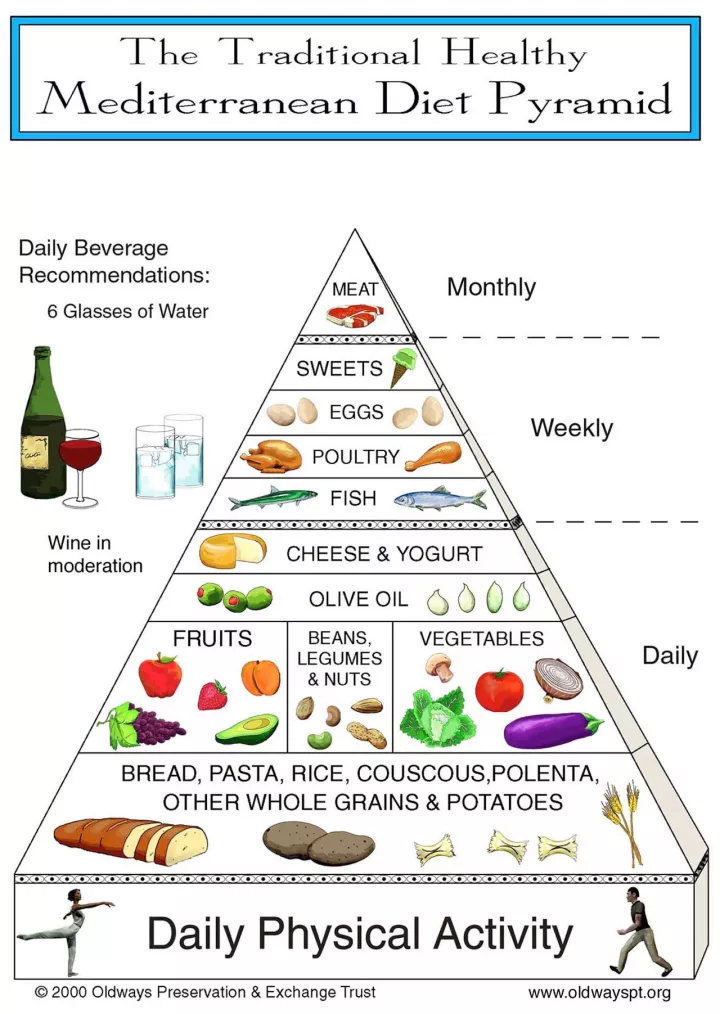
2. 地中海飲食法 – 4.1/5
源自地中海沿岸國家的飲食智慧,被證實有助延年益壽,並降低癌症與心血管疾病風險。
特色:
- 大量攝取蔬果、橄欖油、豆類與全穀物
- 以橄欖油作為主要脂肪來源
專家觀點:營養結構完整且食材選擇多元,是維持體態的理想選擇。
3. 彈性素食法 – 3.9/5
融合素食主義與彈性飲食概念,允許偶爾攝取肉類的植物性飲食法。
特色:
- 以素食為主,每週僅少量攝取肉類(如1次)
- 提倡從豆類、豆腐等植物性食材獲取蛋白質
專家觀點:比嚴格素食更易執行,既能保證營養均衡,又能實現循序漸進的體重管理。
由梅奧診所研發,此飲食法旨在建立終生健康的飲食習慣。
特色:
- 基於梅奧診所健康體重金字塔
- 優先選擇新鮮蔬果、全穀物、瘦肉蛋白與不飽和脂肪
- 提倡飲食調整搭配體能活動
專家觀點: 對控制糖尿病與促進長期健康特別有效。

5. TLC飲食法 – 3.9/5
治療性生活方式改變(TLC)飲食法專為透過健康飲食降低膽固醇設計。
特色:
- 減少飽和脂肪攝取
- 鼓勵食用瘦肉、家禽、魚類、蔬果與全穀物
專家觀點: 有益心臟健康且無明顯缺點的飲食方案。

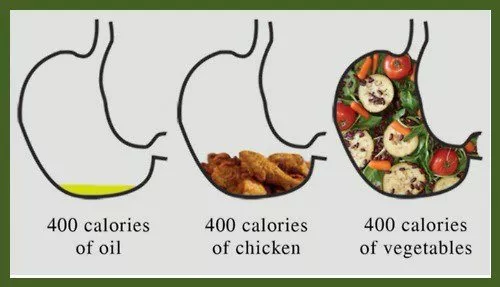
6. 體積飲食法 – 3.8/5
此飲食法專注食物密度,優先選擇低卡高飽足感食物如蔬果。
特色:
- 高含水量食物提升飽足感
- 限制披薩、餅乾等高熱量密度食物
專家觀點: 對心臟健康與糖尿病預防效果顯著。
7. 長壽飲食法 – 2.9/5
此飲食法源自禪宗思想,透過食物追求陰陽平衡。
特色:
- 強調有機全穀物、當地時令蔬菜與植物性蛋白
- 減少加工食品、肉類與乳製品攝取
專家觀點: 難以長期維持且缺乏科學實證支持健康主張。

8. 間歇性斷食法 – 2.5/5
又稱5:2飲食法,透過間歇性禁食減少熱量攝取。
特點:
- 五天正常飲食,兩天非連續日大幅減少熱量攝取(500-600大卡)。
專家觀點: 短期減重有效,但長期可能導致營養缺乏。

9. 阿特金斯飲食法 – 2.4/5
透過低碳水化合物、高蛋白質飲食改變代謝模式,從燃燒碳水化合物轉為燃燒脂肪。
特點:
- 排除大部分碳水化合物,專注攝取蛋白質與脂肪。
專家觀點: 短期可能有效,但可能增加腎臟負擔並導致認知疲勞。

10. 原始人飲食法 – 2.3/5
靈感源自遠古人類飲食模式,排除現代加工食品。
特點:
- 以瘦肉、魚類、水果和蔬菜為主
- 避免穀物、乳製品和加工食品
專家觀點: 不符合現代生活型態,且碳水化合物攝取量不足。